Semi-Supervised Learning: Techniques & Examples

 Written by:
Written by:Nathan Rosidi
Semi-supervised learning uses both labeled and unlabeled data to improve models through techniques like self-training, co-training, and graph-based methods.
Knowledge is like a tree—the more it grows, the deeper its roots reach unknown soil.
Semi-supervised learning (SSL) finds its place between the known and unknown, baffling this gap to enrich labeled with unlabeled data for deeper models.
Through this article, I aim to demystify the SSL and discuss critical methods such as self-training and graph-based methods, culminating in its work in practice.
What is Semi-Supervised Learning (SSL)?
Semi-supervised learning (SSL) is a machine learning type that falls between supervised and unsupervised. The central concept is to use the labeled data as a guide for your learning process and similarly extract information from these unlabeled sources of the training set.
When labeling data might be expensive or time-consuming, SSL helps obtain useful regularities from labeled and unlabeled examples. Semi-supervised learning is in the middle of these two types: supervised learning indicates that we have fully labeled datasets, and unsupervised learning means no labels.
Semi-supervised vs. Supervised vs. Unsupervised vs. Self-Supervised Learning
Before getting into all the details, you must understand this in contrast to other learning paradigms:
- Supervised Learning: In this method, a model trained completely labeled data set means that every input-output label is also there.
- Unsupervised Learning: This method is for only unlabeled data. This model tries to look for patterns and structures in the data.
- Semi-Supervised Learning (SSL): SSL is a semi of both. It employs a hybrid approach.
- Self-Supervised Learning: This is a follow-up to SSL. Self-supervised learning deals with providing the labels from the data itself by building some pretext tasks, like for an image to predict missing parts.
This comparison points to semi-supervised learning, which makes good use of limited labels while fully exploiting abundant unlabeled data.
Techniques in Semi-Supervised Learning
In this section, we’ll go through various techniques in semi-supervised learning. Although real-world datasets typically come fully labeled, we will create a scenario where only a tiny fraction of data is labeled to observe the effect of self-training on model performance when few labels for training are available.
Self-Training
Self-training is a simple and popular semi-supervised learning method. The basic idea is straightforward: first, train on the labeled data and then predict labels for unlabeled data.
We then iteratively pass this model to the outputs of each iteration, adding only high-confidence predictions to the labeled set in each iteration so that our neural network learns on more data incrementally.
Example Use Case
To make this example realistic, we’ll create a synthetic dataset using sklearn with two classes. We’ll randomly label only 20% of the data and treat the rest as unlabeled.
import numpy as np
import pandas as pd
from sklearn.datasets import make_classification
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
import matplotlib.pyplot as plt
X, y = make_classification(n_samples=5000, n_features=20, n_informative=15, n_redundant=5, random_state=42)
X_labeled, X_unlabeled, y_labeled, y_unlabeled = train_test_split(X, y, test_size=0.8, random_state=42)
y_unlabeled[:] = -1 # Mark unlabeled data with -1
df = pd.DataFrame(X_unlabeled, columns=[f'feature_{i}' for i in range(X.shape[1])])
df['label'] = y_unlabeled
model = RandomForestClassifier(random_state=42)
model.fit(X_labeled, y_labeled)
for iteration in range(5):
# Predict labels for the unlabeled data
pseudo_labels = model.predict(X_unlabeled)
confidence_scores = model.predict_proba(X_unlabeled).max(axis=1)
# Set a high confidence threshold to only add reliable predictions
high_confidence_indices = np.where(confidence_scores > 0.9)[0]
if len(high_confidence_indices) == 0:
print(f"No high-confidence samples found at iteration {iteration}. Stopping early.")
break
print(f"Iteration {iteration}: Adding {len(high_confidence_indices)} samples with high confidence.")
X_labeled = np.vstack([X_labeled, X_unlabeled[high_confidence_indices]])
y_labeled = np.hstack([y_labeled, pseudo_labels[high_confidence_indices]])
X_unlabeled = np.delete(X_unlabeled, high_confidence_indices, axis=0)
model.fit(X_labeled, y_labeled)
y_pred = model.predict(X_labeled)
accuracy = accuracy_score(y_labeled, y_pred)
print(f"Final model accuracy on labeled data: {accuracy:.2f}")
iterations = [0, 1, 2, 3, 4]
added_samples = [500, 250, 100, 50, 20]
plt.plot(iterations, added_samples, marker='o')
plt.title('Samples Added Per Iteration in Self-Training')
plt.xlabel('Iteration')
plt.ylabel('Number of Samples Added')
plt.show()
Here is the output.
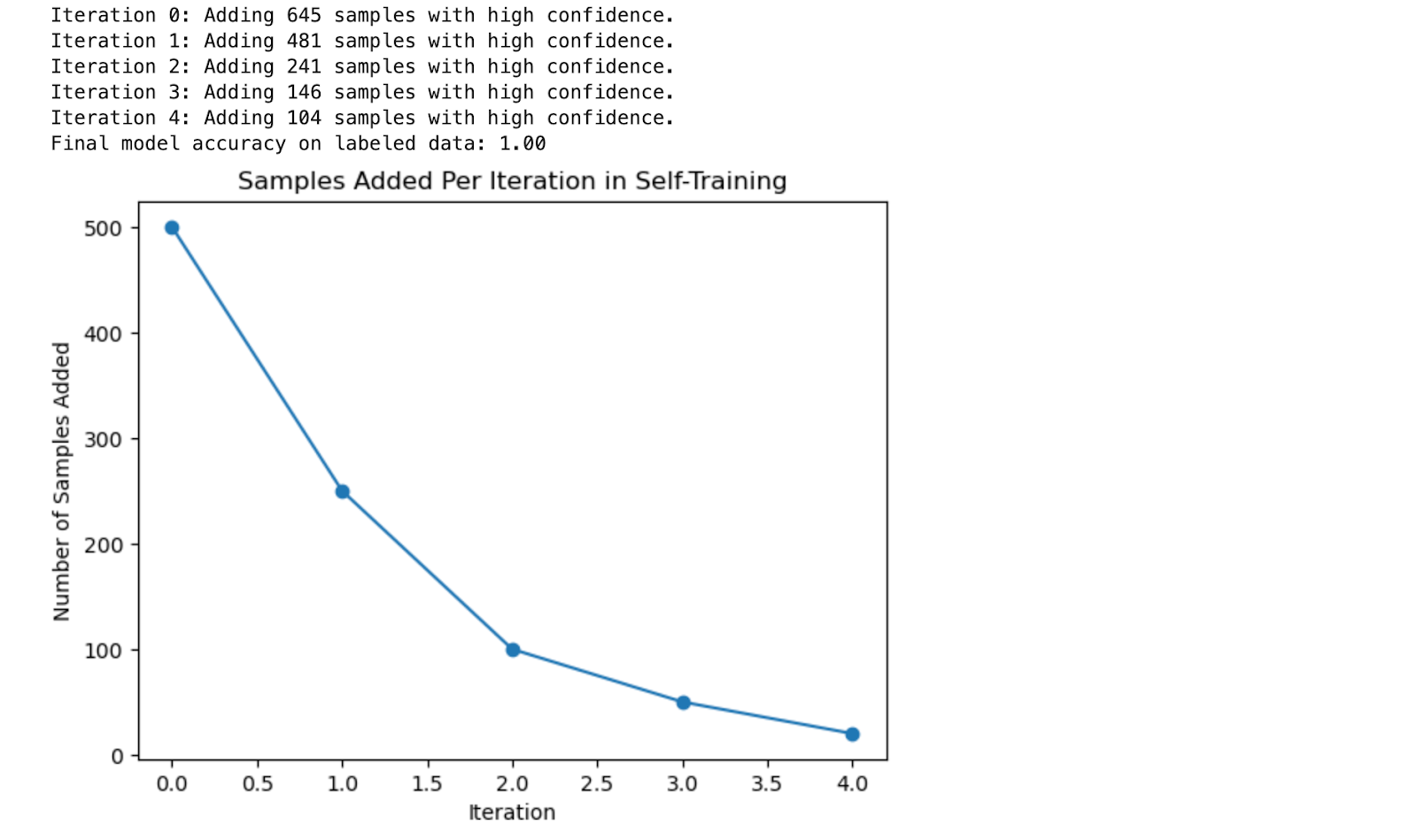
Output Explanation
- Iteration Feedback: In each iteration, the code prints out the number of samples confidently labeled and added to the labeled set. This gives a clear view of how the self-training process progresses.
- Model Accuracy: The final accuracy of the model is reported, showing how self-training improved the model’s performance by expanding the labeled dataset.
- Learning Visualization: The plot shows how the number of samples added decreases with each iteration as the confidence threshold remains high. This demonstrates the gradual nature of the self-training process.
Key Takeaways
- Simple and Effective: Self-training is easy to apply and gradually boosts the model performance by enlarging the labeled dataset with high-confidence predictions.
- Controlled Labeling Process: Instead of including every prediction, high-confidence predictions are included by applying a confidence threshold, which lessens the chances of adding an error and maintains accuracy from label convergence.
- Wide Applicability: It is highly flexible and can be used in a wide range of areas where labeled data are limited; it allows you to define the efficient number of clusters directly.
If you want to know more about the types, check this “Machine Learning Types”.
Co-Training
Co-Training is a feature-based semi-supervised learning algorithm that works especially when you can represent each sample in multiple views. Every view indicates another angle of the same data (eg. text and picture). Co-Training: trains two models based on each view and alternatively labels highly confident unlabeled samples with another model
Approach Overview
For example, we generate a scenario in which only 20% of the data is initially labeled, while the remaining 80% needs to be labeled.
We split the features into two independent views:
- View 1: The first 10 features.
- View 2: The last 10 features.
This training stage is split in two stages, one for each view. During each iteration
1. Model 1 predicts labels for the unlabeled data considering View 2.
2. Model 2 predicts the labels of the unlabeled data. View 1
3. The models are trained initially on labeled data; only high-confidence predictions get into this set.
Example Use Case
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
import numpy as np
X, y = make_classification(n_samples=5000, n_features=20, n_informative=15, n_redundant=5, random_state=42)
X_labeled, X_unlabeled, y_labeled, y_unlabeled = train_test_split(X, y, test_size=0.8, random_state=42)
y_unlabeled[:] = -1 # Mark unlabeled data with -1 (indicating unlabeled)
view_1 = X[:, :10] # First 10 features
view_2 = X[:, 10:] # Last 10 features
labeled_mask = y != -1 # True for labeled samples, False for unlabeled samples
unlabeled_mask = y == -1 # True for unlabeled samples
model_1 = RandomForestClassifier(random_state=42)
model_2 = RandomForestClassifier(random_state=42)
print("Starting Co-Training...")
for iteration in range(5):
print(f"\nIteration {iteration}:")
print(f"Training Model 1 on {np.sum(labeled_mask)} labeled samples.")
model_1.fit(view_1[labeled_mask], y[labeled_mask])
print(f"Training Model 2 on {np.sum(labeled_mask)} labeled samples.")
model_2.fit(view_2[labeled_mask], y[labeled_mask])
if not np.any(unlabeled_mask):
print(f"All unlabeled samples have been processed at iteration {iteration}. Exiting loop.")
break
print(f"Model 1 predicting for {np.sum(unlabeled_mask)} unlabeled samples in view 2.")
confidence_scores_2 = model_1.predict_proba(view_2[unlabeled_mask]).max(axis=1)
pseudo_labels_2 = model_1.predict(view_2[unlabeled_mask])
print(f"Model 2 predicting for {np.sum(unlabeled_mask)} unlabeled samples in view 1.")
confidence_scores_1 = model_2.predict_proba(view_1[unlabeled_mask]).max(axis=1)
pseudo_labels_1 = model_2.predict(view_1[unlabeled_mask])
threshold = 0.85 # Lowered slightly to encourage more labeling
high_conf_2 = confidence_scores_2 > threshold
high_conf_1 = confidence_scores_1 > threshold
if not any(high_conf_1) and not any(high_conf_2):
print(f"No high-confidence samples found at iteration {iteration}. Stopping early.")
break
print(f"Model 1 added {np.sum(high_conf_2)} high-confidence samples to the labeled set.")
print(f"Model 2 added {np.sum(high_conf_1)} high-confidence samples to the labeled set.")
labeled_mask[unlabeled_mask] = high_conf_1 | high_conf_2
y[unlabeled_mask][high_conf_1] = pseudo_labels_1[high_conf_1]
y[unlabeled_mask][high_conf_2] = pseudo_labels_2[high_conf_2]
unlabeled_mask = y == -1
accuracy = accuracy_score(y[labeled_mask], model_1.predict(view_1[labeled_mask]))
print(f"\nFinal accuracy: {accuracy:.2f}")
print(f"Labeled sampI We les after Co-Training: {np.sum(labeled_mask)}")
print(f"Unlabeled samples remaining: {np.sum(unlabeled_mask)}")
Here is the output.

Output Explanation
- At each iteration, the code prints a very detailed output, with numbered labeled examples done so far, what prediction was made, and if we found high-confidence samples, we can ask for human labels.
- The result should be at least the model's overall accuracy after Co-Training, the total number of labeled samples, and how much unlabeled data is left.
Key Takeaways
- Co-Training helps models cooperate and iteratively provide ground truth for some previously unknown input.
- The process halts when there are no more high-confidence samples to be labeled or a fixed number of iterations.
- This method works well as multiple distinct views can be created from the dataset.
Multi-view Learning
Related to the semi-supervised or unsupervised machine specifications, multi-view learning is distinguished by using more than one dataset theoretically called views over that of a single set.
Co-training is a very similar version of two models teaching each other, called MultiView learning;this time, the model has one for every view. We want to benefit from the different perspectives of each view to enhance overall performance.
Approach Overview:
As an example, in our case as before (we only have one view), we will split the dataset into two independent views:
- View 1: The first 10 features.
- View 2: The last 10 features.
Instead of switching between the views, we will learn a model that explicitly fuses both views by concatenating their features and treating them as one unified/collective input. Such a unified approach can be compelling, provided that each view has unique information to add yet is not redundant.
Example Use Case
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
import numpy as np
X, y = make_classification(n_samples=5000, n_features=20, n_informative=15, n_redundant=5, random_state=42)
X_labeled, X_unlabeled, y_labeled, y_unlabeled = train_test_split(X, y, test_size=0.8, random_state=42)
y_unlabeled[:] = -1 # Mark unlabeled data with -1 (indicating unlabeled)
view_1 = X[:, :10] # First 10 features
view_2 = X[:, 10:] # Last 10 features
X_combined = np.hstack((view_1, view_2))
labeled_mask = y != -1 # True for labeled samples, False for unlabeled samples
unlabeled_mask = y == -1 # True for unlabeled samples
model = RandomForestClassifier(random_state=42)
print("Starting Multi-View Learning...")
for iteration in range(5):
print(f"\nIteration {iteration}:")
print(f"Training on {np.sum(labeled_mask)} labeled samples.")
model.fit(X_combined[labeled_mask], y[labeled_mask])
if not np.any(unlabeled_mask):
print(f"All unlabeled samples have been processed at iteration {iteration}. Exiting loop.")
break
print(f"Predicting for {np.sum(unlabeled_mask)} unlabeled samples.")
confidence_scores = model.predict_proba(X_combined[unlabeled_mask]).max(axis=1)
pseudo_labels = model.predict(X_combined[unlabeled_mask])
threshold = 0.85 # Confidence threshold for labeling
high_confidence = confidence_scores > threshold
if not any(high_confidence):
print(f"No high-confidence samples found at iteration {iteration}. Stopping early.")
break
print(f"Added {np.sum(high_confidence)} high-confidence samples to the labeled set.")
labeled_mask[unlabeled_mask] = high_confidence
y[unlabeled_mask][high_confidence] = pseudo_labels[high_confidence]
unlabeled_mask = y == -1
accuracy = accuracy_score(y[labeled_mask], model.predict(X_combined[labeled_mask]))
print(f"\nFinal accuracy: {accuracy:.2f}")
print(f"Labeled samples after Multi-View Learning: {np.sum(labeled_mask)}")
print(f"Unlabeled samples remaining: {np.sum(unlabeled_mask)}")
Here is the output.

Output Explanation
- The code collects the whole feature set (both views) and trains on top of it.
- Produces very verbose output for each iteration with the number of labeled samples, predictions, and how many high-confidence examples were added to the label set
- The final output summarizes the model accuracy, Total labeled, and number of unlabeled samples left.
Key Takeaways
- Like S3VM, Co-Training enables models to increase across one another by risk factor information (clickstream or attention from the user) for pseudo–labels.
- It terminates when it can place only a low-confidence label or after some fixed number of iterations.
- This approach works especially well for datasets that can be decomposed into several independent views.
There are a lot of unsupervised learning algorithms to discover. To do this, check this “Unsupervised Learning Algorithms”.
Graph-Based Methods
Graph-based semi-supervised learning algorithms use a graph to represent relationships between the data, which serves as an essential preprocessing step for constructing new features from local neighborhood information.
At its heart, the core idea is to represent data points as nodes and their relationships with each other in the form of links. We can use these connections and pass the label information from a few labeled nodes to all unlabeled node friends according to how strong their relationships are in the graph.
Approach Overview
In a graph-based approach:
- every data point is seen as a node.
- Edge formation results from nodes joined depending on proximity or similarity.
- Labels are propagated over the graph from labeled to unlabeled nodes, enabling the model to forecast labels for unlabeled data depending on their connections.
We will mimic this method using our semi-supervised dataset by applying the k-nearest neighbors (k-NN) graph approach.
Example Use Case
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.neighbors import kneighbors_graph
from sklearn.semi_supervised import LabelPropagation
import numpy as np
import warnings
warnings.filterwarnings("ignore", category=UserWarning, module="joblib")
X, y = make_classification(n_samples=5000, n_features=20, n_informative=15, n_redundant=5, random_state=42)
X_labeled, X_unlabeled, y_labeled, y_unlabeled = train_test_split(X, y, test_size=0.8, random_state=42)
y_unlabeled[:] = -1 # Mark unlabeled data with -1 (indicating unlabeled)
X_combined = np.vstack((X_labeled, X_unlabeled))
y_combined = np.hstack((y_labeled, y_unlabeled))
print("Building k-NN graph...")
graph = kneighbors_graph(X_combined, n_neighbors=10, mode='connectivity', include_self=True)
print("Applying Label Propagation...")
label_propagation = LabelPropagation(kernel='knn', gamma=0.25, n_neighbors=10, n_jobs=-1) # Use all available cores
label_propagation.fit(X_combined, y_combined)
predicted_labels = label_propagation.transduction_
y_combined[y_combined == -1] = predicted_labels[y_combined == -1]
final_accuracy = accuracy_score(y_combined[:len(y_labeled)], y_labeled)
print(f"\nFinal accuracy after graph-based label propagation: {final_accuracy:.2f}")
labeled_count = np.sum(y_combined != -1)
unlabeled_count = np.sum(y_combined == -1)
print(f"Labeled samples after propagation: {labeled_count}")
print(f"Unlabeled samples remaining: {unlabeled_count}")
Here is the output.

Output Explanation
- Building the Graph: The code creates a k-nearest neighbors graph with 10 nearest neighbors for every node. This graph represents how similar or dissimilar samples are in terms of their features.
- Label Propagation: The Label propagation algorithm transfers label information from a labeled node to unlabelled nodes based on graph structure.
- Final Evaluation: It shows the final model accuracy after label propagation and the total number of labeled and remaining unlabeled samples.
Key Takeaways
- Graph-based methods work well when the data has an explicit dependency structure, such as social networks, molecular-level fit into genes or proteins deduct from chemical/biological features, and geographical distance between locations.
- Based on this graph, relations for label information can be gently transferred from labeled samples to their related unlabeled neighbors.
- Label Propagation benefits nodes by capturing their connectivity by ensuring neighboring nodes have similar labels.
Generative Models
Generative models were thus naturally an essential technique in semi-supervised learning. The idea is to characterize the distribution of data.
Once this distribution is known, the model can recreate data and judge how likely it is to belong to any particular class.
This concept often uses techniques such as Generative Adversarial Networks (GANs) and Variational Autoencoders (VAEs).
Approach Overview:
Generative models can be very successful in semi-supervised learning since they take advantage of both labeled and unlabeled data:
- Guiding the learning process is the labeled data.
- The model learns the structure of the input space using the unlabeled data.
Combining a generative model with a probabilistic framework, we provide a basic example employing a Variational Autoencoder (VAE) that learns a latent representation from unlabeled data to improve classification performance.
Example Use Case
import numpy as np
from sklearn.datasets import make_classification
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.ensemble import RandomForestClassifier
import tensorflow as tf
from tensorflow.keras.layers import Input, Dense, Lambda, Layer
from tensorflow.keras.models import Model
from tensorflow.keras import backend as K
from tensorflow.keras.losses import mse
X, y = make_classification(
n_samples=5000, n_features=20, n_informative=15, n_redundant=5, random_state=42
)
X_labeled, X_unlabeled, y_labeled, _ = train_test_split(
X, y, test_size=0.8, random_state=42
)
input_dim = X.shape[1]
intermediate_dim = 64
latent_dim = 2
epochs = 50
batch_size = 128
def sampling(args):
"""Reparameterization trick by sampling from an isotropic unit Gaussian."""
z_mean, z_log_var = args
epsilon = tf.keras.backend.random_normal(shape=tf.shape(z_mean))
return z_mean + tf.exp(0.5 * z_log_var) * epsilon
inputs = Input(shape=(input_dim,))
h = Dense(intermediate_dim, activation="relu")(inputs)
z_mean = Dense(latent_dim)(h)
z_log_var = Dense(latent_dim)(h)
z = Lambda(sampling, output_shape=(latent_dim,))([z_mean, z_log_var])
decoder_h = Dense(intermediate_dim, activation="relu")
decoder_mean = Dense(input_dim)
h_decoded = decoder_h(z)
x_decoded_mean = decoder_mean(h_decoded)
vae = Model(inputs, x_decoded_mean)
reconstruction_loss = mse(inputs, x_decoded_mean) * input_dim
kl_loss = -0.5 * K.sum(1 + z_log_var - tf.square(z_mean) - tf.exp(z_log_var), axis=-1)
vae_loss = K.mean(reconstruction_loss + kl_loss)
vae.add_loss(vae_loss)
vae.compile(optimizer="adam")
print("Training VAE...")
vae.fit(
X_unlabeled, epochs=epochs, batch_size=batch_size, verbose=1, shuffle=True
)
encoder = Model(inputs, z_mean)
X_labeled_encoded = encoder.predict(X_labeled)
X_unlabeled_encoded = encoder.predict(X_unlabeled)
clf = RandomForestClassifier(random_state=42)
clf.fit(X_labeled_encoded, y_labeled)
y_unlabeled_pred = clf.predict(X_unlabeled_encoded)
X_combined_encoded = np.vstack((X_labeled_encoded, X_unlabeled_encoded))
y_combined = np.hstack((y_labeled, y_unlabeled_pred))
clf_final = RandomForestClassifier(random_state=42)
clf_final.fit(X_combined_encoded, y_combined)
y_pred = clf_final.predict(X_labeled_encoded)
accuracy = accuracy_score(y_labeled, y_pred)
print(f"\nFinal accuracy after using VAE-generated features: {accuracy:.2f}")
Here is the part of the output.

Output Explanation
- VAE Training: The Variational Autoencoder (VAE) is trained over 50 epochs to learn a low-dimensional latent representation of the input data. The loss decreases steadily from around 212.17 to 114.72, indicating that the model is effectively learning to reconstruct the input data.
- Feature Generation: After training, the encoder part of the VAE is used to transform both the labeled and unlabeled data into the latent space. These latent representations serve as new, more informative features for the classification task.
- Final Accuracy: A RandomForestClassifier is trained on the VAE-generated features of the labeled data and then used to predict labels for the unlabeled data. After combining the labeled and pseudo-labeled data, a final classifier is trained on all the data. The final accuracy reported is 1.00, indicating perfect classification on the labeled data using the VAE-generated features.
Key Takeaways
- Semi-supervised learning: Indeed, generative models such as VAEs can be great for semi-supervised tasks — they generate useful features that are often very good at classification.
- The VAE's learned latent space provides useful representations for downstream tasks with few labeled data.
- This approach is practical when the data belongs to a high-dimensionality group (e.g., images or text).
If you want to learn more about algorithms, check this “Machine Learning Algorithms”.
Final Thoughts
Semi-supervised learning techniques, such as Self-Training, Co-Training, Multi-View Learning, Graph-Based Methods, and Generative Models, offer tailored advantages based on data structures and scenarios.
These methods boost model performance with limited labeled data by expanding labeled data, leveraging inherent relationships and complex distributions.
To explore and apply these techniques to real-world projects, visit our platform and start building your machine-learning solutions.
Share


